Abstract

Pancreatic cancer, a deadly malignancy with a median survival of less than 1 year, urgently requires biomarkers for prognosis prediction. MPZL2, associated with poor outcomes in other cancers, was analyzed in pancreatic cancer using TCGA and GTEX data. Our study found increased MPZL2 expression in PAAD, which can distinguish patients from healthy individuals with an AUC of 0.919. High MPZL2 levels correlate with worse survival outcomes and identify malignant tumor cells as the primary source. CMap analysis suggests clofibrate may reverse MPZL2 - driven molecular changes. In conclusion, MPZL2 is a promising prognostic biomarker and therapeutic target in PAAD.
Keywords: MPZL2, Pancreatic cancer, TCGA
Dear Editor
Pancreatic cancer, a malignancy challenging to diagnose and treat, ranks as the third - leading cause of cancer - related mortality. Since 2000, the median survival duration of this disease has remained less than 1 year [1] . It is emerging as an increasingly prevalent cause of cancer - related mortality. When the cancer is in its confined stage, patients often present with advanced - stage disease owing to the absence or ambiguity of symptoms [2] . Therefore, there is an urgent need to identify a biomarker capable of more precisely predicting the prognosis and immunotherapy effectiveness in pancreatic cancer patients.
MPZL2 has been demonstrated to have a close association with multiple types of cancers. MPZL2, expressed in lymphoid organs, the thymus, and other epithelial structures, is capable of maintaining the stem-like properties of glioblastoma [3] It has been reported that elevated MPZL2 expression in acute myeloid leukemia and hepatocellular carcinoma is linked to adverse prognosis and recurrence [4,5] . However, the expression of MPZL2 and its implications in pancreatic cancer have never been clarified.
In this study, we primarily utilized the pancreatic cancer dataset from TCGA and the normal sample data from the GTEX database to perform in - depth bioinformatics analyses of MPZL2. The proteomic data of MPZL2 were sourced from the CPTAC database. We evaluated the prognostic and diagnostic implications using Kaplan - Meier survival curves, COX regression models, and ROC curves. Single - cell and spatial transcriptomic analyses determined the cell populations that exhibited significant MPZL2 expression. Additionally, the eXtreme Sum (XSum) method was employed for CMap analysis to identify drugs sensitive to patients with high MPZL2 expression.
Our findings demonstrate that MPZL2 exhibits increased mRNA and protein levels in PAAD (Figure 1A-B). It can accurately distinguish between the pancreatic cancer patient group and the normal group, with an AUC of 0.919 (Figure 1C). Simultaneously, we discovered that elevated MPZL2 expression was significantly associated with poorer overall survival (OS), disease - specific survival (DSS), disease - free interval (DFI), and progression - free interval (PFI) (Figure 1D). Both univariate and multivariate Cox analyses indicated that MPZL2 was detrimental to the overall survival of PAAD patients (Figure 1E) and served as an independent risk factor for PAAD. CMap analysis revealed that clofibrate might reverse the molecular signatures resulting from MPZL2 expression dysregulation, thus counteracting the tumor - promoting effects mediated by MPZL2 (Figure 1F). Single - cell and spatial transcriptomic analyses demonstrated that malignant tumor cells were the specific cell types where MPZL2 was located (Figure 1G-J), and the content of these cells was positively correlated with MPZL2 expression (Figure 1K). Furthermore, spatial transcriptomic analysis revealed that the average expression of MPZL2 in the tumor core and tumor boundary regions was significantly higher than that in the normal regions, and this difference was statistically significant (Figure 1L).
Figure 1 Expression and Prognostic Significance of MPZL2 in Pancreatic Cancer. (A - B) MPZL2 expression in normal and tumor tissues. (C) Diagnostic efficiency of the ROC curve of MPZL2 based on TCGA data; (D) Survival curves of MPZL2 in the TCGA dataset; (E) Univariate and multivariate analyses of MPZL2; (G) Potential small - molecule compounds and drugs predicted by the XSum algorithm to reverse the biological effects induced by MPZL2 gene - expression dysregulation; (G - J) Cellular localization of MPZL2. (K) Spearman's correlation between MPZL2 expression and microenvironmental components at the spatial - transcriptomic resolution. (L) Differences in MPZL2 expression levels among malignant regions (Mal), tumor - boundary regions (Bdy), and normal regions (nMal) at the spatial - transcriptomic resolution.

In conclusion, our study highlights MPZL2 as a valuable prognostic biomarker and a potential therapeutic target in PAAD, bearing great significance for guiding clinical drug development.
Abbreviations
Not applicable.
Supplementary Material
Supplementary methods, results, spectra, figures.
Acknowledgements
We are very grateful to TCGA, GTEX,GEO and CPTAC database for data support. In addition, we thank Photothermal Biology (https://grswsci.top/) for helping with visualization.
Author contributions
ZZ and YK was responsible for writing and authoring the article, ZZ,LJ and YF processed the data, and HC were in charge of reviewing the article.
Ethics approval and consent to participate
The research did not involve any human participants or animals, and therefore did not require approval from an ethics committee. All data used in this study were obtained from publicly available sources and were analyzed in accordance with ethical guidelines and regulations.
Funding information
Not applicable.
Competing Interests
The authors declare that they have no existing or potential commercial or financial relationships that could create a conflict of interest at the time of conducting this study.
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
References
[1] Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12-49. https://doi.org/10.3322/caac.21820
[2] Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008-20. https://doi.org/10.1016/S0140-6736(20)30974-0
[3] Ohtsu N, Nakatani Y, Yamashita D, Ohue S, Ohnishi T, Kondo T. Eva1 Maintains the Stem-like Character of Glioblastoma-Initiating Cells by Activating the Noncanonical NF-kappaB Signaling Pathway. Cancer Res. 2016;76(1):171-81. https://doi.org/10.1158/0008-5472.CAN-15-0884
[4] Ni Q, Chen Z, Zheng Q, Xie D, Li JJ, Cheng S, et al. Epithelial V-like antigen 1 promotes hepatocellular carcinoma growth and metastasis via the ERBB-PI3K-AKT pathway. Cancer Sci. 2020;111(5):1500-13. https://doi.org/10.1111/cas.14331
[5] Yu P, Lan H, Song X, Pan Z. High Expression of the SH3TC2-DT/SH3TC2 Gene Pair Associated With FLT3 Mutation and Poor Survival in Acute Myeloid Leukemia: An Integrated TCGA Analysis. Front Oncol. 2020;10:829. https://doi.org/10.3389/fonc.2020.00829
 Figures
Figures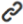 References
References Peer
Peer Information
InformationFigure 1 Expression and Prognostic Significance of MPZL2 in Pancreatic Cancer. (A - B) MPZL2 expression in normal and tumor tissues. (C) Diagnostic efficiency of the ROC curve of MPZL2 based on TCGA data; (D) Survival curves of MPZL2 in the TCGA dataset; (E) Univariate and multivariate analyses of MPZL2; (G) Potential small - molecule compounds and drugs predicted by the XSum algorithm to reverse the biological effects induced by MPZL2 gene - expression dysregulation; (G - J) Cellular localization of MPZL2. (K) Spearman's correlation between MPZL2 expression and microenvironmental components at the spatial - transcriptomic resolution. (L) Differences in MPZL2 expression levels among malignant regions (Mal), tumor - boundary regions (Bdy), and normal regions (nMal) at the spatial - transcriptomic resolution.

[1] Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12-49. https://doi.org/10.3322/caac.21820
[2] Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008-20. https://doi.org/10.1016/S0140-6736(20)30974-0
[3] Ohtsu N, Nakatani Y, Yamashita D, Ohue S, Ohnishi T, Kondo T. Eva1 Maintains the Stem-like Character of Glioblastoma-Initiating Cells by Activating the Noncanonical NF-kappaB Signaling Pathway. Cancer Res. 2016;76(1):171-81. https://doi.org/10.1158/0008-5472.CAN-15-0884
[4] Ni Q, Chen Z, Zheng Q, Xie D, Li JJ, Cheng S, et al. Epithelial V-like antigen 1 promotes hepatocellular carcinoma growth and metastasis via the ERBB-PI3K-AKT pathway. Cancer Sci. 2020;111(5):1500-13. https://doi.org/10.1111/cas.14331
[5] Yu P, Lan H, Song X, Pan Z. High Expression of the SH3TC2-DT/SH3TC2 Gene Pair Associated With FLT3 Mutation and Poor Survival in Acute Myeloid Leukemia: An Integrated TCGA Analysis. Front Oncol. 2020;10:829. https://doi.org/10.3389/fonc.2020.00829
Peer-review Terminology
Identity transparency: Single anonymized
Reviewer interacts with: Editor
Review information published:
Review reports
Reviewer identities if reviewer opts in
Author/reviewer communication
Details
© 2025 The Author(s). Life Conflux published by Life Conflux Press Limited on behalf of Conflux Science.
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.
Publication History
Received 2024-10-18
Accepted 2024-12-21
Published 2024-12-30


