Vol. 2 No. 1
Full Issue
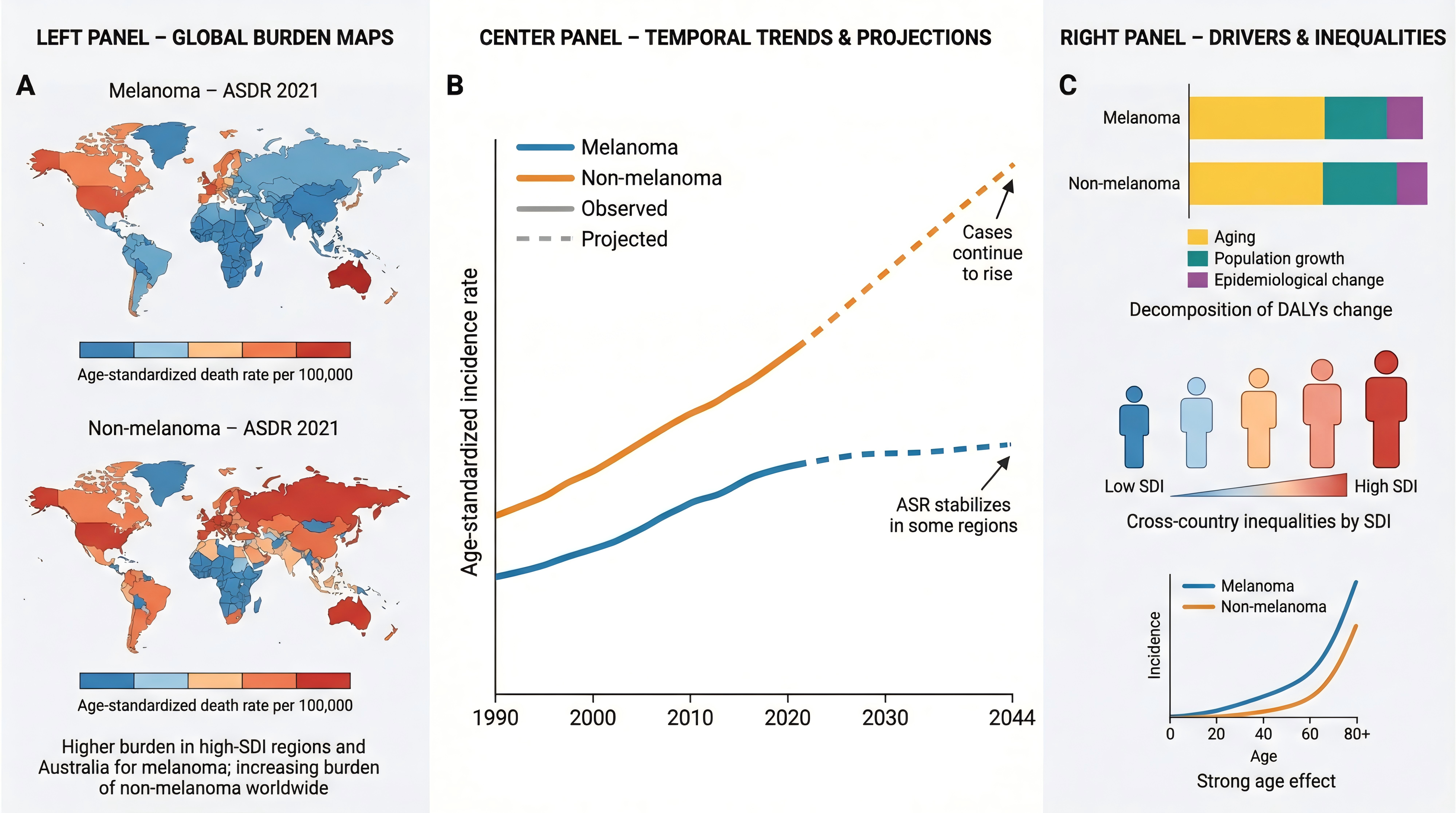
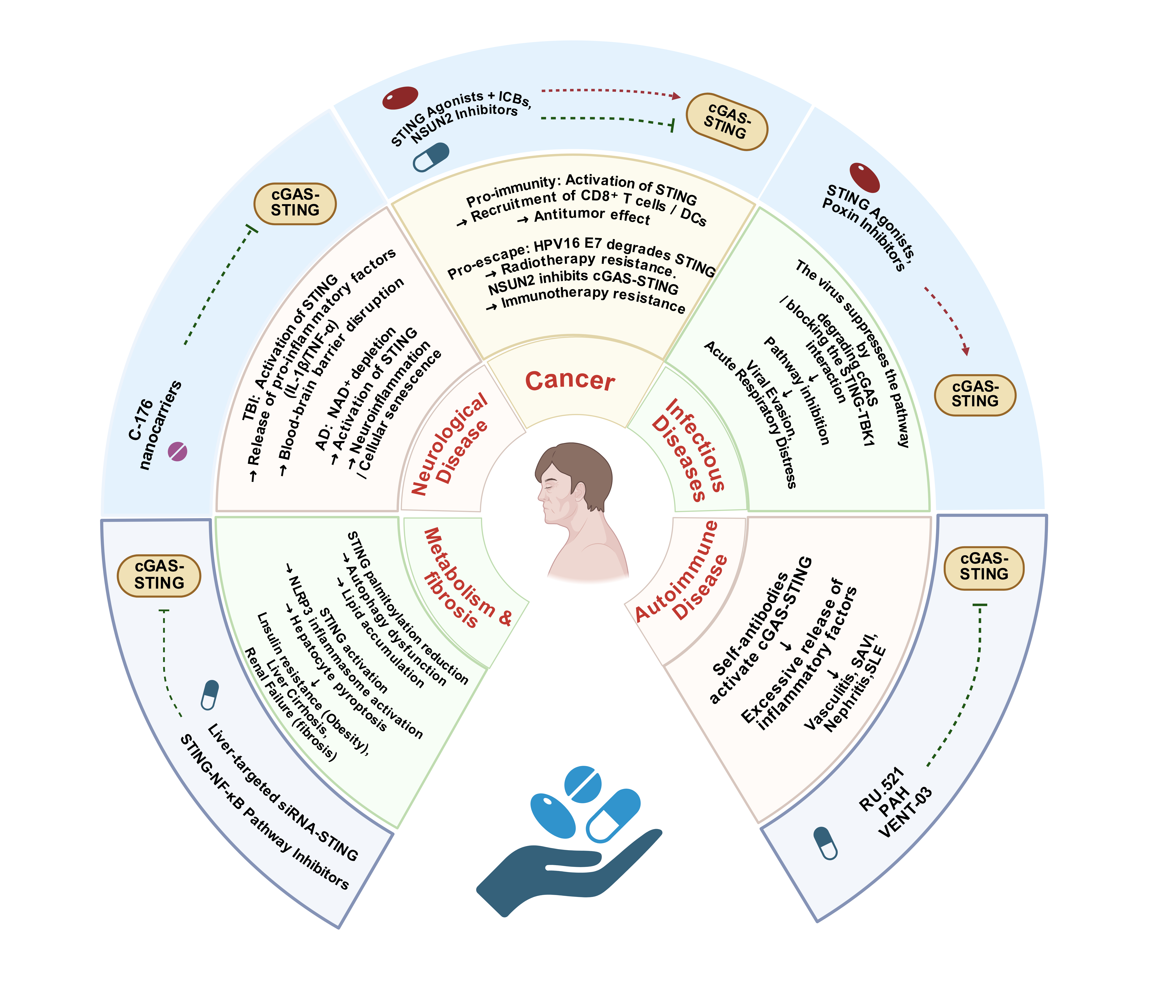
Biomedical Data Science and Analytics
Clinical Decision Support and AI in Medicine
Digital Health and Public Health Informatics
Medical Informatics Cases, Reviews and Applications
Journal Information
- Journal: Life Conflux
- Online ISSN: 3078-4816
- Publisher: Life Conflux Press Limited
- Peer Review: Single-anonymous
- Publication Model: Open Access
- Publishing Frequency: Quarterly
- Languages: English
- DOI Prefix: 10.71321

Submit Your Research
Fast-track peer review process

Journal Metrics
Impact factors & citation analysis
Research Areas
And many more specialized fields within medical informatics, digital health, and healthcare technology
Open Access Benefits
Life Conflux promotes open science principles, ensuring that groundbreaking research in medical informatics and digital health reaches the global scientific community without barriers, accelerating scientific discovery and innovation in healthcare technology and clinical informatics.
Creative Commons License

Life Conflux operates under a Creative Commons Attribution 4.0 International License, enabling unrestricted use, distribution, and reproduction while ensuring proper attribution to original authors and research integrity.
Copyright & Author Rights
Authors retain full copyright ownership of their work while granting Life Conflux the right to publish and distribute. All submissions must comply with our rigorous ethical standards and originality requirements. Published content becomes freely accessible to the global research community, promoting scientific advancement and knowledge sharing.
Key Requirements: Original research, ethical compliance, proper citations, and agreement to publication terms.
Publication Ethics
Life Conflux maintains the highest standards of publication ethics, preventing plagiarism, ensuring research integrity, and upholding international best practices. We implement comprehensive screening processes and follow COPE (Committee on Publication Ethics) guidelines to maintain scientific credibility.